2011/08/16
Alternate Models of RNA Looping on Nova-1 Protein in Splicing
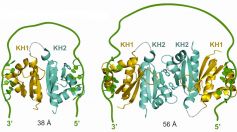
Nova proteins are implicated in alternate splicing regulation of specific neuronal genes and expressed exclusively in neurons. Recent issue of journal STRUCTURE (1) reported the results of structural study of Nova-1 that summarized the efforts of eleven co-authors from eight different institutions to understand how the structure works in alternate splicing. Researchers from United States (MSKCC, Rockefeller University, Scripps Research Institute, Cornell University and Harvard University) and Spain (CIC bioGUNE) had to apply a powerful arsenal of modern research techniques, including X-ray crystallography, NMR, Isothermal titration calorimetry, Ultracentrifuge, Filter Binding Assays, and many others, to derive two alternate models of possible RNA Looping on the surface of Nova-1 (Figure) that can explain Nova ability either enhance or inhibit exon inclusion, thereby potentially explaining Nova's functional role in splicing regulation. The three first authors of the publication in Structure, Marianna Teplova (MSKCC, USA), Lucy Malinina (CIC bioGUNE, Spain), and Jennifer C. Darnell (Rockefeller University, USA) contributed equally to this exhausting study.
Reference:(1) Teplova M, Malinina L, Darnell JC, Song J, Lu M, Abagyan R, Musunuru K, Teplov A, Burley SK, Darnell RB, Patel DJ (2011) Protein-RNA and Protein-Protein Recognition by Dual KH1/2 Domains of the Neuronal Splicing Factor Nova-1. Structure; Jul 13; 19(7):930-44.
See a large version of the first picture





