2012/02/27
The structure of the dimerization domain of the tumor suppressor ING4 reveals the functional organization of the ING family of chromatin binding proteins.
A collaborative work between the groups of Ignacio Palmero (CSIC), Guillermo Montoya (CNIO) and Francisco J Blanco (CIC bioGUNE) shows that the dimerization domain of ING4 forms an antiparallel coiled-coil structure with important implications for chromatin binding. Dimerization is essential for the ING4 mediated apoptosis induction in response to genotoxic stress and for the inhibition of anchorage independent cell growth.
Together with our previous findings on the structural organization of ING4, these results suggests that ING4 can bind simultaneously two histone H3 tail on the same or different nucleosomes.
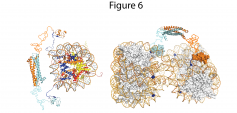
Figure: Model of the ING4 dimer engaging the histone H3 N-terminal tails of the same nucleosome through its PHD fingers. The flexible nuclear localization signal regions of ING4 and the histone H3 tails are represented as curvy lines. The nucleosome structure is represented with the DNA as an orange rod (backbone) and blue-green sticks (bases) and the histone proteins a ribbons of different colors: H3 in blue, H4 in red, H2A in magenta and H2B in yellow.
See a large version of the first picture





