2014/07/18
Using in vivo-biotinylated ubiquitin to describe a mitotic exit ubiquitome from human cells.
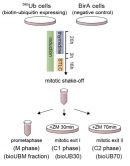
During mitotic cell division chromosomes need to be repackaged into daughter cells, a complicated process guided by several protein regulators, which are themselves tightly regulated. Targeted destruction of those proteins by cellular machinery is achieved by first modifying them with a small 'tag' called ubiquitin. The lab of Ikerbasque Research Professor Ugo Mayor in bioGUNE has developed a unique technique to identify ubiquitinated proteins in a number of systems, and after applying it to flies (Drosophila) as well as to mice in recent papers, has now used this system in collaboration with coworkers at the University of Cambridge (UK) to investigate the regulation of mitosis in human cells.
Coupling purification with mass spectrometry, a series of mitotic regulators that are targeted for polyubiquitination in mitotic exit have been identified in this work. Some of those are new substrates of the Anaphase Promoting Complex/Cyclosome (APC/C), with KIFC1 and RacGAP1/Cyk4 being validated as two such targets involved respectively in timely mitotic spindle disassembly and cell spreading. In vivo biotin-tagging of ubiquitin can provide valuable information about the role of ubiquitin-mediated regulation in processes required for rebuilding interphase cells.
This study was recently published in the journal MCP
Min M, Mayor U, Dittmar G, Lindon C
See a large version of the first picture





